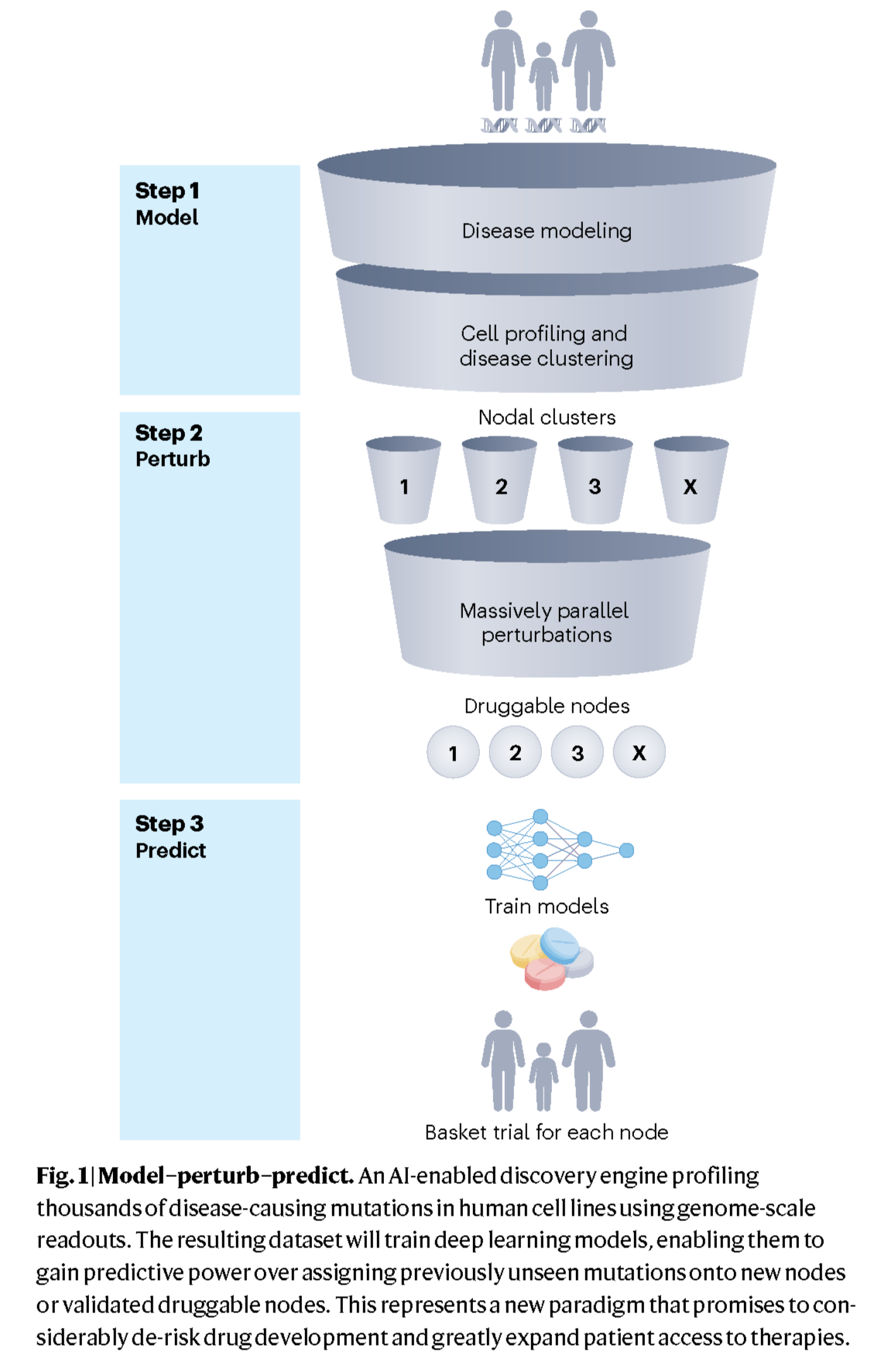
A roadmap for finding druggable nodes
In a comment in Nature Medicine, Jillian Shaw and Anna Greka describe a framework for identifying druggable “nodes” shared by clinically distinct genetic diseases in an effort to accelerate the development of treatments that target these nodes. In this Comment, the team lays out a three-step method for discovering these targets: (1) build cellular models of thousands of disease-causing genetic variants in a pooled fashion; (2) conduct massively parallel perturbation screens using single-cell and high-content readouts to find shared mechanisms underlying thousands of mutations; (3) use these datasets to train AI models to predict druggable nodes. These insights may lead to the grouping of patients according to their molecular profiles rather than their traditional diagnoses.